|
The research group of Prof. Wei Xie at School of Life Sciences, Tsinghua University published an article paper in Nature Genetics on December 5th, entitled “Dynamic epigenomic landscapes during early lineage specification in mouse embryos”. This study reported the dynamics of epigenomic landscapes during early lineage specification in mouse.
In mammals, all somatic development originates from lineage segregation in early embryos. Notably, early cell fate commitment is accompanied by extensive epigenetic reprogramming. For example, DNA methylation shows drastic demethylation and remethylation during early embryogenesis. Work from Wei Xie’s lab and other groups also showed the majority of histone modifications from parents are erased prior to implantation. How the zygotic epigenome is established and facilitates the lineage specification is a fundamental question in developmental biology. However, due to the limited availability of materials and the difficulty of tissue isolation in early embryos, lineage-specific establishment of epigenomes in peri- and postipmlantation embryos is poorly characterized.
To address this question, Wei Xie’s group reported comprehensive investigation of transcriptomes and base-resolution methylomes for early lineages in peri- and postimplantation mouse embryos. They found allele- and lineage-specific de novo methylation at CG and CH sites, leading to differential methylation between embryonic and extraembryonic lineages at promoters of lineage regulators, gene bodies, and DNA methylation valleys. By determining chromatin architecture using Hi-C across the same developmental period, they showed both global demethylation and remethylation in early development correlate with chromatin compartments. Dynamic local methylation is evident during gastrulation, which revealed maps of putative regulatory elements. Finally, they found that de novo methylation patterning does not strictly require implantation. These data unveiled dynamic transcriptomes, DNA methylomes, and 3D chromatin landscapes during the earliest mammalian lineage specification.
Investigator Wei Xie from School of Life Sciences at Tsinghua University is the corresponding author of this work. Ph.D candidates Yu Zhang, Yunlong Xiang, Zhenhai Du and Research Associate Qiangzong Yin from School of Life Sciences at Tsinghua University are the first authors of this work. Feng Xu and Xu Peng from the Institute of Molecular and Cell Biology at A*STAR in Singapore made great contributions to this work. Other collaborators include Lei Li’s group from Institute of Zoology at Chinese Academy of Sciences, Jianlong Wang’s group from Icahn School of Medicine at Mount Sinai, New York in the United States. This work is supported by the funding provided by the National Key R&D Program of China, National Basic Research Program of China (973 program), the National Natural Science Foundation of China, the National Basic Research Program of China, the funding from the THU-PKU Center for Life Sciences, the Youth Thousand Scholar Program of China, Beijing Advanced Innovation Center for Structural Biology, the Biomedical Research Council of A*STAR (Agency for Science, Technology and Research) at Singapore, National Institute of Health (NIH), and Howard Hughes Medical Institute.
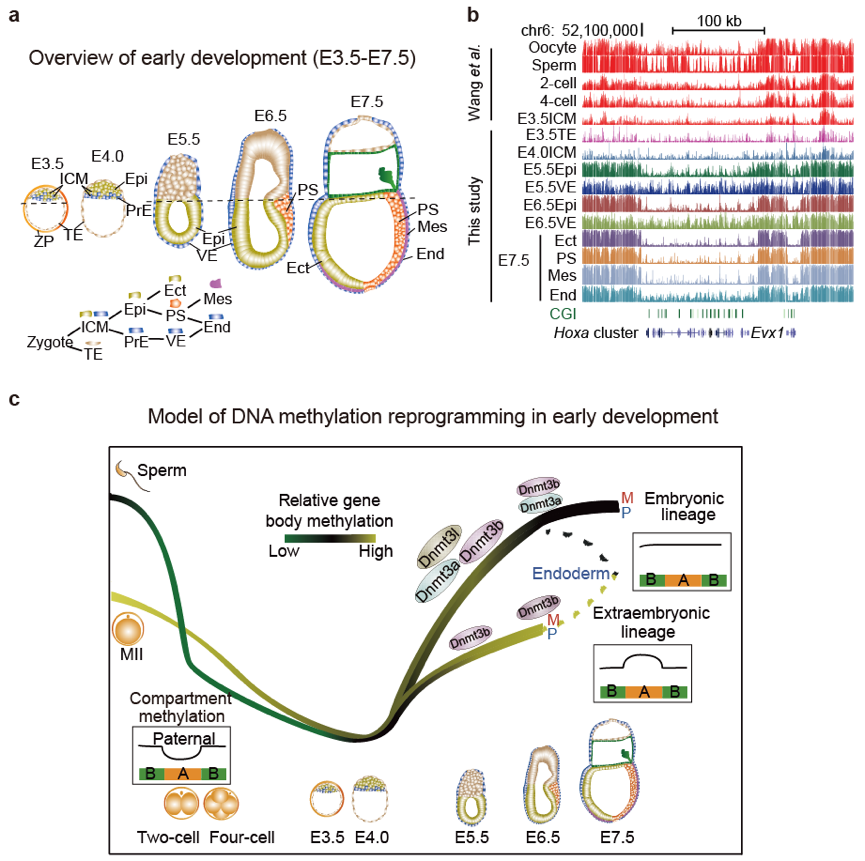
Figure 1. a, Overview of mouse embryonic development from E3.5 to E7.5. b, Lineage-specific DNA methylation landscapes in mouse embryos. c, A model of allele and lineage-specific DNA methylation reprogramming in mouse early embryos.
Paper link:
https://www.nature.com/articles/s41588-017-0003-x
|
